May. 10, 2024
Faculty from the George W. Woodruff School of Mechanical Engineering, including Associate Professors Gregory Sawicki and Aaron Young, have been awarded a five-year, $2.6 million Research Project Grant (R01) from the National Institutes of Health (NIH).
“We are grateful to our NIH sponsor for this award to improve treatment of post-stroke individuals using advanced robotic solutions,” said Young, who is also affiliated with Georgia Tech's Neuro Next Initiative.
The R01 will support a project focused on using optimization and artificial intelligence to personalize exoskeleton assistance for individuals with symptoms resulting from stroke. Sawicki and Young will collaborate with researchers from the Emory Rehabilitation Hospital including Associate Professor Trisha Kesar.
“As a stroke researcher, I am eagerly looking forward to making progress on this project, and paving the way for leading-edge technologies and technology-driven treatment strategies that maximize functional independence and quality of life of people with neuro-pathologies," said Kesar.
The intervention for study participants will include a training therapy program that will use biofeedback to increase the efficiency of exosuits for wearers.
Kinsey Herrin, senior research scientist in the Woodruff School and Neuro Next Initiative affiliate, explained the extended benefits of the study, including being able to increase safety for stroke patients who are moving outdoors. “One aspect of this project is testing our technologies on stroke survivors as they're walking outside. Being outside is a small thing that many of us take for granted, but a devastating loss for many following a stroke.”
Sawicki, who is also an associate professor in the School of Biological Sciences and core faculty in Georgia Tech's Institute for Robotics and Intelligent Machines, is also looking forward to the project. "This new project is truly a tour de force that leverages a highly talented interdisciplinary team of engineers, clinical scientists, and prosthetics/orthotics experts who all bring key elements needed to build assistive technology that can work in real-world scenarios."
News Contact
Chloe Arrington
Communications Officer II
George W. Woodruff School of Mechanical Engineering
May. 06, 2024
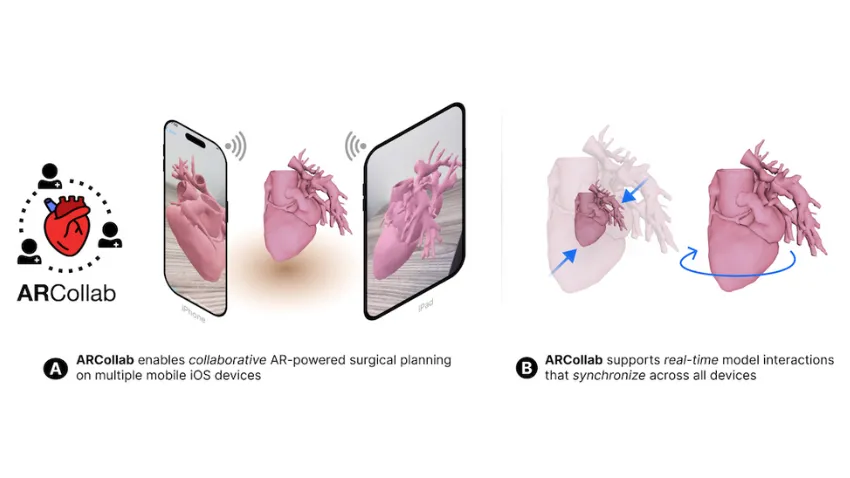
Cardiologists and surgeons could soon have a new mobile augmented reality (AR) tool to improve collaboration in surgical planning.
ARCollab is an iOS AR application designed for doctors to interact with patient-specific 3D heart models in a shared environment. It is the first surgical planning tool that uses multi-user mobile AR in iOS.
The application’s collaborative feature overcomes limitations in traditional surgical modeling and planning methods. This offers patients better, personalized care from doctors who plan and collaborate with the tool.
Georgia Tech researchers partnered with Children’s Healthcare of Atlanta (CHOA) in ARCollab’s development. Pratham Mehta, a computer science major, led the group’s research.
“We have conducted two trips to CHOA for usability evaluations with cardiologists and surgeons. The overall feedback from ARCollab users has been positive,” Mehta said.
“They all enjoyed experimenting with it and collaborating with other users. They also felt like it had the potential to be useful in surgical planning.”
ARCollab’s collaborative environment is the tool’s most novel feature. It allows surgical teams to study and plan together in a virtual workspace, regardless of location.
ARCollab supports a toolbox of features for doctors to inspect and interact with their patients' AR heart models. With a few finger gestures, users can scale and rotate, “slice” into the model, and modify a slicing plane to view omnidirectional cross-sections of the heart.
Developing ARCollab on iOS works twofold. This streamlines deployment and accessibility by making it available on the iOS App Store and Apple devices. Building ARCollab on Apple’s peer-to-peer network framework ensures the functionality of the AR components. It also lessens the learning curve, especially for experienced AR users.
ARCollab overcomes traditional surgical planning practices of using physical heart models. Producing physical models is time-consuming, resource-intensive, and irreversible compared to digital models. It is also difficult for surgical teams to plan together since they are limited to studying a single physical model.
Digital and AR modeling is growing as an alternative to physical models. CardiacAR is one such tool the group has already created.
However, digital platforms lack multi-user features essential for surgical teams to collaborate during planning. ARCollab’s multi-user workspace progresses the technology’s potential as a mass replacement for physical modeling.
“Over the past year and a half, we have been working on incorporating collaboration into our prior work with CardiacAR,” Mehta said.
“This involved completely changing the codebase, rebuilding the entire app and its features from the ground up in a newer AR framework that was better suited for collaboration and future development.”
Its interactive and visualization features, along with its novelty and innovation, led the Conference on Human Factors in Computing Systems (CHI 2024) to accept ARCollab for presentation. The conference occurs May 11-16 in Honolulu.
CHI is considered the most prestigious conference for human-computer interaction and one of the top-ranked conferences in computer science.
M.S. student Harsha Karanth and alumnus Alex Yang (CS 2022, M.S. CS 2023) co-authored the paper with Mehta. They study under Polo Chau, an associate professor in the School of Computational Science and Engineering.
The Georgia Tech group partnered with Timothy Slesnick and Fawwaz Shaw from CHOA on ARCollab’s development.
“Working with the doctors and having them test out versions of our application and give us feedback has been the most important part of the collaboration with CHOA,” Mehta said.
“These medical professionals are experts in their field. We want to make sure to have features that they want and need, and that would make their job easier.”
News Contact
Bryant Wine, Communications Officer
bryant.wine@cc.gatech.edu
Mar. 19, 2024
Computer science educators will soon gain valuable insights from computational epidemiology courses, like one offered at Georgia Tech.
B. Aditya Prakash is part of a research group that will host a workshop on how topics from computational epidemiology can enhance computer science classes.
These lessons would produce computer science graduates with improved skills in data science, modeling, simulation, artificial intelligence (AI), and machine learning (ML).
Because epidemics transcend the sphere of public health, these topics would groom computer scientists versed in issues from social, financial, and political domains.
The group’s virtual workshop takes place on March 20 at the technical symposium for the Special Interest Group on Computer Science Education (SIGCSE). SIGCSE is one of 38 special interest groups of the Association for Computing Machinery (ACM). ACM is the world’s largest scientific and educational computing society.
“We decided to do a tutorial at SIGCSE because we believe that computational epidemiology concepts would be very useful in general computer science courses,” said Prakash, an associate professor in the School of Computational Science and Engineering (CSE).
“We want to give an introduction to concepts, like what computational epidemiology is, and how topics, such as algorithms and simulations, can be integrated into computer science courses.”
Prakash kicks off the workshop with an overview of computational epidemiology. He will use examples from his CSE 8803: Data Science for Epidemiology course to introduce basic concepts.
This overview includes a survey of models used to describe behavior of diseases. Models serve as foundations that run simulations, ultimately testing hypotheses and making predictions regarding disease spread and impact.
Prakash will explain the different kinds of models used in epidemiology, such as traditional mechanistic models and more recent ML and AI based models.
Prakash’s discussion includes modeling used in recent epidemics like Covid-19, Zika, H1N1 bird flu, and Ebola. He will also cover examples from the 19th and 20th centuries to illustrate how epidemiology has advanced using data science and computation.
“I strongly believe that data and computation have a very important role to play in the future of epidemiology and public health is computational,” Prakash said.
“My course and these workshops give that viewpoint, and provide a broad framework of data science and computational thinking that can be useful.”
While humankind has studied disease transmission for millennia, computational epidemiology is a new approach to understanding how diseases can spread throughout communities.
The Covid-19 pandemic helped bring computational epidemiology to the forefront of public awareness. This exposure has led to greater demand for further application from computer science education.
Prakash joins Baltazar Espinoza and Natarajan Meghanathan in the workshop presentation. Espinoza is a research assistant professor at the University of Virginia. Meghanathan is a professor at Jackson State University.
The group is connected through Global Pervasive Computational Epidemiology (GPCE). GPCE is a partnership of 13 institutions aimed at advancing computational foundations, engineering principles, and technologies of computational epidemiology.
The National Science Foundation (NSF) supports GPCE through the Expeditions in Computing program. Prakash himself is principal investigator of other NSF-funded grants in which material from these projects appear in his workshop presentation.
[Related: Researchers to Lead Paradigm Shift in Pandemic Prevention with NSF Grant]
Outreach and broadening participation in computing are tenets of Prakash and GPCE because of how widely epidemics can reach. The SIGCSE workshop is one way that the group employs educational programs to train the next generation of scientists around the globe.
“Algorithms, machine learning, and other topics are fundamental graduate and undergraduate computer science courses nowadays,” Prakash said.
“Using examples like projects, homework questions, and data sets, we want to show that the topics and ideas from computational epidemiology help students see a future where they apply their computer science education to pressing, real world challenges.”
News Contact
Bryant Wine, Communications Officer
bryant.wine@cc.gatech.edu
Jan. 29, 2024
For over three decades, a highly accurate early diagnostic test for ovarian cancer has eluded physicians. Now, scientists in the Georgia Tech Integrated Cancer Research Center (ICRC) have combined machine learning with information on blood metabolites to develop a new test able to detect ovarian cancer with 93 percent accuracy among samples from the team’s study group.
John McDonald, professor emeritus in the School of Biological Sciences, founding director of the ICRC, and the study’s corresponding author, explains that the new test’s accuracy is better in detecting ovarian cancer than existing tests for women clinically classified as normal, with a particular improvement in detecting early-stage ovarian disease in that cohort.
The team’s results and methodologies are detailed in a new paper, “A Personalized Probabilistic Approach to Ovarian Cancer Diagnostics,” published in the March 2024 online issue of the medical journal Gynecologic Oncology. Based on their computer models, the researchers have developed what they believe will be a more clinically useful approach to ovarian cancer diagnosis — whereby a patient’s individual metabolic profile can be used to assign a more accurate probability of the presence or absence of the disease.
“This personalized, probabilistic approach to cancer diagnostics is more clinically informative and accurate than traditional binary (yes/no) tests,” McDonald says. “It represents a promising new direction in the early detection of ovarian cancer, and perhaps other cancers as well.”
The study co-authors also include Dongjo Ban, a Bioinformatics Ph.D. student in McDonald’s lab; Research Scientists Stephen N. Housley, Lilya V. Matyunina, and L.DeEtte (Walker) McDonald; Regents’ Professor Jeffrey Skolnick, who also serves as Mary and Maisie Gibson Chair in the School of Biological Sciences and Georgia Research Alliance Eminent Scholar in Computational Systems Biology; and two collaborating physicians: University of North Carolina Professor Victoria L. Bae-Jump and Ovarian Cancer Institute of Atlanta Founder and Chief Executive Officer Benedict B. Benigno. Members of the research team are forming a startup to transfer and commercialize the technology, and plan to seek requisite trials and FDA approval for the test.
Silent killer
Ovarian cancer is often referred to as the silent killer because the disease is typically asymptomatic when it first arises — and is usually not detected until later stages of development, when it is difficult to treat.
McDonald explains that while the average five-year survival rate for late-stage ovarian cancer patients, even after treatment, is around 31 percent — but that if ovarian cancer is detected and treated early, the average five-year survival rate is more than 90 percent.
“Clearly, there is a tremendous need for an accurate early diagnostic test for this insidious disease,” McDonald says.
And although development of an early detection test for ovarian cancer has been vigorously pursued for more than three decades, the development of early, accurate diagnostic tests has proven elusive. Because cancer begins on the molecular level, McDonald explains, there are multiple possible pathways capable of leading to even the same cancer type.
“Because of this high-level molecular heterogeneity among patients, the identification of a single universal diagnostic biomarker of ovarian cancer has not been possible,” McDonald says. “For this reason, we opted to use a branch of artificial intelligence — machine learning — to develop an alternative probabilistic approach to the challenge of ovarian cancer diagnostics.”
Metabolic profiles
Georgia Tech co-author Dongjo Ban, whose thesis research contributed to the study, explains that “because end-point changes on the metabolic level are known to be reflective of underlying changes operating collectively on multiple molecular levels, we chose metabolic profiles as the backbone of our analysis.”
“The set of human metabolites is a collective measure of the health of cells,” adds coauthor Jeffrey Skolnick, “and by not arbitrarily choosing any subset in advance, one lets the artificial intelligence figure out which are the key players for a given individual.”
Mass spectrometry can identify the presence of metabolites in the blood by detecting their mass and charge signatures. However, Ban says, the precise chemical makeup of a metabolite requires much more extensive characterization.
Ban explains that because the precise chemical composition of less than seven percent of the metabolites circulating in human blood have, thus far, been chemically characterized, it is currently impossible to accurately pinpoint the specific molecular processes contributing to an individual's metabolic profile.
However, the research team recognized that, even without knowing the precise chemical make-up of each individual metabolite, the mere presence of different metabolites in the blood of different individuals, as detected by mass spectrometry, can be incorporated as features in the building of accurate machine learning-based predictive models (similar to the use of individual facial features in the building of facial pattern recognition algorithms).
“Thousands of metabolites are known to be circulating in the human bloodstream, and they can be readily and accurately detected by mass spectrometry and combined with machine learning to establish an accurate ovarian cancer diagnostic,” Ban says.
A new probabilistic approach
The researchers developed their integrative approach by combining metabolomic profiles and machine learning-based classifiers to establish a diagnostic test with 93 percent accuracy when tested on 564 women from Georgia, North Carolina, Philadelphia and Western Canada. 431 of the study participants were active ovarian cancer patients, and while the remaining 133 women in the study did not have ovarian cancer.
Further studies have been initiated to study the possibility that the test is able to detect very early-stage disease in women displaying no clinical symptoms, McDonald says.
McDonald anticipates a clinical future where a person with a metabolic profile that falls within a score range that makes cancer highly unlikely would only require yearly monitoring. But someone with a metabolic score that lies in a range where a majority (say, 90%) have previously been diagnosed with ovarian cancer would likely be monitored more frequently — or perhaps immediately referred for advanced screening.
Citation: https://doi.org/10.1016/j.ygyno.2023.12.030
Funding
This research was funded by the Ovarian Cancer Institute (Atlanta), the Laura Crandall Brown Foundation, the Deborah Nash Endowment Fund, Northside Hospital (Atlanta), and the Mark Light Integrated Cancer Research Student Fellowship.
Disclosure
Study co-authors John McDonald, Stephen N. Housley, Jeffrey Skolnick, and Benedict B. Benigno are the co-founders of MyOncoDx, Inc., formed to support further research, technology transfer, and commercialization for the team’s new clinical tool for the diagnosis of ovarian cancer.
News Contact
Writer: Renay San Miguel
Communications Officer II/Science Writer
College of Sciences
404-894-5209
Editor: Jess Hunt-Ralston
Nov. 29, 2023
The National Institute of Health (NIH) has awarded Yunan Luo a grant for more than $1.8 million to use artificial intelligence (AI) to advance protein research.
New AI models produced through the grant will lead to new methods for the design and discovery of functional proteins. This could yield novel drugs and vaccines, personalized treatments against diseases, and other advances in biomedicine.
“This project provides a new paradigm to analyze proteins’ sequence-structure-function relationships using machine learning approaches,” said Luo, an assistant professor in Georgia Tech’s School of Computational Science and Engineering (CSE).
“We will develop new, ready-to-use computational models for domain scientists, like biologists and chemists. They can use our machine learning tools to guide scientific discovery in their research.”
Luo’s proposal improves on datasets spearheaded by AlphaFold and other recent breakthroughs. His AI algorithms would integrate these datasets and craft new models for practical application.
One of Luo’s goals is to develop machine learning methods that learn statistical representations from the data. This reveals relationships between proteins’ sequence, structure, and function. Scientists then could characterize how sequence and structure determine the function of a protein.
Next, Luo wants to make accurate and interpretable predictions about protein functions. His plan is to create biology-informed deep learning frameworks. These frameworks could make predictions about a protein’s function from knowledge of its sequence and structure. It can also account for variables like mutations.
In the end, Luo would have the data and tools to assist in the discovery of functional proteins. He will use these to build a computational platform of AI models, algorithms, and frameworks that ‘invent’ proteins. The platform figures the sequence and structure necessary to achieve a designed proteins desired functions and characteristics.
“My students play a very important part in this research because they are the driving force behind various aspects of this project at the intersection of computational science and protein biology,” Luo said.
“I think this project provides a unique opportunity to train our students in CSE to learn the real-world challenges facing scientific and engineering problems, and how to integrate computational methods to solve those problems.”
The $1.8 million grant is funded through the Maximizing Investigators’ Research Award (MIRA). The National Institute of General Medical Sciences (NIGMS) manages the MIRA program. NIGMS is one of 27 institutes and centers under NIH.
MIRA is oriented toward launching the research endeavors of young career faculty. The grant provides researchers with more stability and flexibility through five years of funding. This enhances scientific productivity and improves the chances for important breakthroughs.
Luo becomes the second School of CSE faculty to receive the MIRA grant. NIH awarded the grant to Xiuwei Zhang in 2021. Zhang is the J.Z. Liang Early-Career Assistant Professor in the School of CSE.
[Related: Award-winning Computer Models Propel Research in Cellular Differentiation]
“After NIH, of course, I first thanked my students because they laid the groundwork for what we seek to achieve in our grant proposal,” said Luo.
“I would like to thank my colleague, Xiuwei Zhang, for her mentorship in preparing the proposal. I also thank our school chair, Haesun Park, for her help and support while starting my career.”
News Contact
Bryant Wine, Communications Officer
bryant.wine@cc.gatech.edu
Jun. 28, 2023
Alex Robel is improving how computer models of melting ice sheets incorporate data from field expeditions and satellites by creating a new open-access software package — complete with state-of-the-art tools and paired with ice sheet models that anyone can use, even on a laptop or home computer.
Improving these models is critical: while melting ice sheets and glaciers are top contributors to sea level rise, there are still large uncertainties in sea level projections at 2100 and beyond.
“Part of the problem is that the way that many models have been coded in the past has not been conducive to using these kinds of tools,” Robel, an assistant professor in the School of Earth and Atmospheric Sciences, explains. “It's just very labor-intensive to set up these data assimilation tools — it usually involves someone refactoring the code over several years.”
“Our goal is to provide a tool that anyone in the field can use very easily without a lot of labor at the front end,” Robel says. “This project is really focused around developing the computational tools to make it easier for people who use ice sheet models to incorporate or inform them with the widest possible range of measurements from the ground, aircraft and satellites.”
Now, a $780,000 NSF CAREER grant will help him to do so.
The National Science Foundation Faculty Early Career Development Award is a five-year funding mechanism designed to help promising researchers establish a personal foundation for a lifetime of leadership in their field. Known as CAREER awards, the grants are NSF’s most prestigious funding for untenured assistant professors.
“Ultimately,” Robel says, “this project will empower more people in the community to use these models and to use these models together with the observations that they're taking.”
Ice sheets remember
“Largely, what models do right now is they look at one point in time, and they try their best — at that one point in time — to get the model to match some types of observations as closely as possible,” Robel explains. “From there, they let the computer model simulate what it thinks that ice sheet will do in the future.”
In doing so, the models often assume that the ice sheet starts in a state of balance, and that it is neither gaining nor losing ice at the start of the simulation. The problem with this approach is that ice sheets dynamically change, responding to past events — even ones that have happened centuries ago. “We know from models and from decades of theory that the natural response time scale of thick ice sheets is hundreds to thousands of years,” Robel adds.
By informing models with historical records, observations, and measurements, Robel hopes to improve their accuracy. “We have observations being made by satellites, aircraft, and field expeditions,” says Robel. “We also have historical accounts, and can go even further back in time by looking at geological observations or ice cores. These can tell us about the long history of ice sheets and how they've changed over hundreds or thousands of years.”
Robel’s team plans to use a set of techniques called data assimilation to adjust, or ‘nudge’, models. “These data assimilation techniques have been around for a really long time,” Robel explains. “For example, they’re critical to weather forecasting: every weather forecast that you see on your phone was ultimately the product of a weather model that used data assimilation to take many observations and apply them to a model simulation.”
“The next part of the project is going to be incorporating this data assimilation capability into a cloud-based computational ice sheet model,” Robel says. “We are planning to build an open source software package in Python that can use this sort of data assimilation method with any kind of ice sheet model.”
Robel hopes it will expand accessibility. “Currently, it's very labor-intensive to set up these data assimilation tools, and while groups have done it, it usually involves someone re-coding and refactoring the code over several years.”
Building software for accessibility
Robel’s team will then apply their software package to a widely used model, which now has an online, browser-based version. “The reason why that is particularly useful is because the place where this model is running is also one of the largest community repositories for data in our field,” Robel says.
Called Ghub, this relatively new repository is designed to be a community-wide place for sharing data on glaciers and ice sheets. “Since this is also a place where the model is living, by adding this capability to this cloud-based model, we'll be able to directly use the data that's already living in the same place that the model is,” Robel explains.
Users won’t need to download data, or have a high-speed computer to access and use the data or model. Researchers collecting data will be able to upload their data to the repository, and immediately see the impact of their observations on future ice sheet melt simulations. Field researchers could use the model to optimize their long-term research plans by seeing where collecting new data might be most critical for refining predictions.
“We really think that it is critical for everyone who's doing modeling of ice sheets to be doing this transient data simulation to make sure that our simulations across the field are all doing the best possible job to reproduce and match observations,” Robel says. While in the past, the time and labor involved in setting up the tools has been a barrier, “developing this particular tool will allow us to bring transient data assimilation to essentially the whole field.”
Bringing Real Data to Georgia’s K-12 Classrooms
The broad applications and user-base expands beyond the scientific community, and Robel is already developing a K-12 curriculum on sea level rise, in partnership with Georgia Tech CEISMC Researcher Jayma Koval. “The students analyze data from real tide gauges and use them to learn about statistics, while also learning about sea level rise using real data,” he explains.
Because the curriculum matches with state standards, teachers can download the curriculum, which is available for free online in partnership with the Southeast Coastal Ocean Observing Regional Association (SECOORA), and incorporate it into their preexisting lesson plans. “We worked with SECOORA to pilot a middle school curriculum in Atlanta and Savannah, and one of the things that we saw was that there are a lot of teachers outside of middle school who are requesting and downloading the curriculum because they want to teach their students about sea level rise, in particular in coastal areas,” Robel adds.
In Georgia, there is a data science class that exists in many high schools that is part of the computer science standards for the state. “Now, we are partnering with a high school teacher to develop a second standards-aligned curriculum that is meant to be taught ideally in a data science class, computer class or statistics class,” Robel says. “It can be taught as a module within that class and it will be the more advanced version of the middle school sea level curriculum.”
The curriculum will guide students through using data analysis tools and coding in order to analyze real sea level data sets, while learning the science behind what causes variations and sea level, what causes sea level rise, and how to predict sea level changes.
“That gets students to think about computational modeling and how computational modeling is an important part of their lives, whether it's to get a weather forecast or play a computer game,” Robel adds. “Our goal is to get students to imagine how all these things are combined, while thinking about the way that we project future sea level rise.”
Jun. 16, 2023
The discovery of nucleic acids is a recent event in the history of scientific phenomenon, and there is still much learn from the enigma that is genetic code.
Advances in computing techniques though have ushered in a new age of understanding the macromolecules that form life as we know it. Now, one Georgia Tech research group is receiving well-deserved accolades for their applications in data science and machine learning toward single-cell omics research.
Students studying under Xiuwei Zhang, an assistant professor in the School of Computational Science and Engineering (CSE), received awards in April at the Atlanta Workshop on Single-cell Omics (AWSOM 2023).
School of CSE Ph.D. student Ziqi Zhang received the best oral presentation award, while Mihir Birfna, an undergraduate student majoring in computer science, took the best poster prize.
Along with providing computational tools for biological researchers, the group’s papers presented at AWSOM 2023 could benefit populations of people as the research could lead to improved disease detection and prevention. They can also provide a better understanding of causes and treatments of cancer and new ability to accurately simulate cellular processes.
“I am extremely proud of the entire research group and very thankful of their work and our teamwork within our lab,” said Xiuwei Zhang. “These awards are encouraging because it means we are on the right track of developing something that will contribute both to the biology community and the computational community.”
Ziqi Zhang presented the group’s findings of their deep learning framework called scDisInFact, which can carry out multiple key single cell RNA-sequencing (scRNA-seq) tasks all at once and outperform current models that focus on the same tasks individually.
The group successfully tested scDisInFact on simulated and real Covid-19 datasets, demonstrating applicability in future studies of other diseases.
Bafna’s poster introduced CLARIFY, a tool that connects biochemical signals occurring within a cell and intercellular communication molecules. Previously, the inter- and intra-cell signaling were often studied separately due to the complexity of each problem.
Oncology is one field that stands to benefit from CLARIFY. CLARIFY helps to understand the interactions between tumor cells and immune cells in cancer microenvironments, which is crucial for enabling success of cancer immunotherapy.
At AWSOM 2023, the group presented a third paper on scMultiSim. This simulator generates data found in multi-modal single-cell experiments through modeling various biological factors underlying the generated data. It enables quantitative evaluations of a wide range of computational methods in single-cell genomics. That has been a challenging problem due to lack of ground truth information in biology, Xiuwei Zhang said.
“We want to answer certain basic questions in biology, like how did we get these different cell types like skin cells, bone cells, and blood cells,” she said. “If we understand how things work in normal and healthy cells, and compare that to the data of diseased cells, then we can find the key differences of those two and locate the genes, proteins, and other molecules that cause problems.”
Xiuwei Zhang’s group specializes in applying machine learning and optimization skills in analysis of single-cell omics data and scRNA-seq methods. Their main interest area is studying mechanisms of cellular differentiation— the process when young, immature cells mature and take on functional characteristics.
A growing, yet effective approach to research in molecular biology, scRNA-seq gives insight of existence and behavior of different types of cells. This helps researchers better understand genetic disorders, detect mechanisms that cause tumors and cancer, and develop new treatments, cures, and drugs.
If microenvironments filled with various macromolecules and genetic material are considered datasets, the need for researchers like Xiuwei Zhang and her group is obvious. These massive, complex datasets present both challenges and opportunities for the group experienced in computational and biological research.
Collaborating authors include School of CSE Ph.D. students Hechen Li and Michael Squires, School of Electrical and Computer Engineering Ph.D. student Xinye Zhao, Wallace H. Coulter Department of Biomedical Engineering Associate Professor Peng Qiu, and Xi Chen, an assistant professor at Southern University of Science and Technology in Shenzhen, China.
The group’s presentations at AWSOM 2023 exhibited how their work makes progress in biomedical research, as well as advancing scientific computing methods in data science, machine learning, and simulation.
scDisInFact is an optimization tool that can perform batch effect removal, condition-associated key gene detection, and perturbation, which is made possible by considering major variation factors in the data. Without considering all these factors, current models can only do these tasks individually, but scDisInFact can do each task better and all at the same time.
CLARIFY delves into how cells employ genetic material to communicate internally, using gene regulatory networks (GRNs) and externally, called cell-cell interactions (CCIs). Many computational methods can infer GRNs and inference methods have been proposed for CCIs, but until CLARIFY, no method existed to infer GRNs and CCIs in the same model.
scMultiSim simulations perform closer to real-world conditions than current simulators that model only one or two biological factors. This helps researchers more realistically test their computational methods, which can guide directions for future method development.
Whether they be computer scientists, biologists, or non-academics alike, the advantage of interdisciplinary and collaborative research, like Xiuwei Zhang’s group, is its wide-reaching impact that advances technology to improve the human condition.
“We’re exploring the possibilities that can be realized by advanced computational methods combined with cutting edge biotechnology,” said Xiuwei Zhang. “Since biotechnology keeps evolving very fast and we want to help push this even further by developing computational methods, together we will propel science forward.”
News Contact
Bryant Wine, Communications Officer
bryant.wine@cc.gatech.edu
Apr. 18, 2023
Our world is powered by chemical reactions. From new medicines and biotechnology to sustainable energy solutions developing and understanding the chemical reactions behind innovations is a critical first step in pioneering new advances. And a key part of developing new chemistries is discovering how the rates of those chemical reactions can be accelerated or changed.
For example, even an everyday chemical reaction, like toasting bread, can substantially change in speed and outcome — by increasing the heat, the speed of the reaction increases, toasting the bread faster. Adding another chemical ingredient — like buttering the bread before frying it — also changes the outcome of the reaction: the bread might brown and crisp rather than toast. The lesson? Certain chemical reactions can be accelerated or changed by adding or altering key variables, and understanding those factors is crucial when trying to create the desired reaction (like avoiding burnt toast!).
Chemists currently use quantum chemistry techniques to predict the rates and energies of chemical reactions, but the method is limited: predictions can usually only be made for up to a few hundred atoms. In order to scale the predictions to larger systems, and predict the environmental effects of reactions, a new framework needs to be developed.
Jesse McDaniel (School of Chemistry and Biochemistry) is creating that framework by leveraging computer modeling techniques. Now, a new NSF CAREER grant will help him do so. The National Science Foundation Faculty Early Career Development Award is a five-year grant designed to help promising researchers establish a foundation for a lifetime of leadership in their field. Known as CAREER awards, the grants are NSF’s most prestigious funding for untenured assistant professors.
“I am excited about the CAREER research because we are really focusing on fundamental questions that are central to all of chemistry,” McDaniel says about the project.
Pioneering a new framework
“Chemical reactions are inherently quantum mechanical in nature,” McDaniel explains. “Electrons rearrange as chemical bonds are broken and formed.” While this type of quantum chemistry can allow scientists to predict the rates and energies of different reactions, these predictions are limited to only tens or hundreds of atoms. That’s where McDaniel’s team comes in. They’re developing modeling techniques based on quantum chemistry that could function over multiple scales, using computer models to scale the predictions. They hope this will help predict environmental effects on chemical reaction rates.
By developing modeling techniques that can be applied to reactions at multiple scales, McDaniel aims to expand scientist’s ability to predict and model chemical reactions, and how they interact with their environments. “Our goal is to understand the microscopic mechanisms and intermolecular interactions through which chemical reactions are accelerated within unique solvation environments such as microdroplets, thin films, and heterogenous interfaces,” McDaniel says. He hopes that it will allow for computational modeling of chemical reactions in much larger systems.
Interdisciplinary research
As a theoretical and computational chemist, McDaniel’s chemistry experiments don’t take place in a typical chemistry lab — rather, they take place in a computer lab, where Georgia Tech’s robust computer science and software development community functions as a key resource.
“We run computer simulations on high performance computing clusters,” McDaniel explains. “In this regard, we benefit from the HPC infrastructure at Georgia Tech, including the Partnership for an Advanced Computing Environment (PACE) team, as well as the computational resources provided in the new CODA building.”
“Software is also a critical part of our research,” he continues. “My colleague Professor David Sherrill and his group are lead developers of the Psi4 quantum chemistry software, and this software comprises a core component of our multi-scale modeling efforts.”
In this respect, McDaniel is eager to to involve the next generation of chemists and computer scientists, showcasing the connection between these different fields. McDaniel’s team will partner with regional high school teachers, collaborating to integrate software and data science tools within the high school educational curriculum.
“One thing I like about this project,” McDaniel says, “is that all types of chemists — organic, inorganic, analytical, bio, physical, etc. — care about how chemical reactions happen, and how reactions are influenced by their surroundings.”
News Contact
Written by Selena Langner
Mar. 02, 2023
Ryan Lawler realized early on in her academic career that a scientist with a great idea can potentially change the world.
“But I didn’t realize the role that real estate can play in that,” said Lawler, general manager of BioSpark Labs – the collaborative, shared laboratory environment taking shape at Science Square at Georgia Tech.
Sitting adjacent to the Tech campus and formerly known as Technology Enterprise Park, Science Square is being reactivated and positioned as a life sciences research destination. The 18-acre site is abuzz with new construction, as an urban mixed-use development rises from the property.
Meanwhile, positioned literally on the ground floor of all this activity is BioSpark Labs, located in a former warehouse, fortuitously adjacent to the Global Center for Medical Innovation. It’s one of the newer best-kept secrets in the Georgia Tech research community.
BioSpark exists because the Georgia Tech Real Estate Office, led by Associate Vice President Tony Zivalich, recognized the need of this kind of lab space. Zivalich and his team have overseen the ideation, design, and funding of the facility, partnering with Georgia Advanced Technology Ventures, as well as the Wallace H. Coulter Department of Biomedical Engineering at Georgia Tech and Emory University, and the core facilities of the Petit Institute for Bioengineering and Bioscience.
“We are in the middle of a growing life sciences ecosystem, part of a larger vision in biotech research,” said Lawler, who was hired on to manage the space, bringing to the job a wealth of experience as a former research scientist and lab manager with a background in molecular and synthetic biology.
Researchers’ Advocate
BioSpark was designed to be a launch pad for high-potential entrepreneurs. It provides a fully equipped and professionally operated wet lab, in addition to a clean room, meeting and office space, to its current roster of clients, five life sciences and biotech startup, a number certain to increase – because BioSpark is undergoing a dramatic expansion that will include 11 more labs (shared and private space), an autoclave room, equipment and storage rooms.
“We want to provide the necessary services and support that an early-stage company needs to begin lab operations on day one,” said Lawler, who has put together a facility with $1.7 million in lab equipment. “I understand our clients’ perspective, I understand researchers and their experiments, and their needs, because I have first-hand proficiency in that world. So, I can advocate on their behalf.”
CO2 incubators, a spectrophotometer, a biosafety cabinet, a fume hood, a -80° freezer, an inverted microscope, and the autoclave are among the wide range of apparatus. Plus, a virtual treasure trove of equipment is available to BioSpark clients off-site through the Core Facilities of the Petit Institute for Bioengineering and Bioscience on the Georgia Tech campus.
“One of the unique things about us is, we’re agnostic,” Lawler said. “That is, our startups can come from anywhere. We have companies that have grown out of labs at Georgia State, Alabama State, Emory, and Georgia Tech. And we have interest from entrepreneurs from San Diego, who are considering relocating people from mature biotech markets to our space.”
Ground Floor Companies
Marvin Whiteley wants to help humans win the war against bacteria, and he has a plan, something he’s been cooking up for about 10 years, which has now manifested in his start-up company, SynthBiome, one of the five startups based at BioSpark Labs.
“We can discover a lot of antibiotics in the lab but translating them into the clinic has been a major challenge – antibiotic resistance is the main reason,” said Whiteley, professor in the School of Biological Sciences at Georgia Tech. “Something might work in a test tube easily enough and it might work in a mouse. But the thing is, bacteria know that mice are different - and and so bacteria act differently in mice than in humans.”
SynthBiome was built to help accelerate drug discovery. With that goal in mind, Whiteley and has team set out to develop a better, more effective preclinical model. “We basically learned to let the bacteria tell us what it’s like to be in a human,” Whiteley said. “So, we created a human environment in a test tube.”
Whiteley has said a desire to help people is foundational to his research. He wants to change how successful therapies are made. The same can be said for Dr. Pooja Tiwari, who launched her company, Arnav Biotech, to develop mRNA-based therapeutics and vaccines. Arnav Biotech also serves as a contract researcher and manufacturer, helping other researchers and companies interested in exploring mRNA in their work.
“There are only a handful of people who have deep knowledge of working in mRNA research, and this limits the access to it” said Tiwari, a former postdoctoral researcher at Georgia Tech and Emory. “We’d like to democratize access to mRNA-based therapeutics and vaccines by developing accessible and cost-effective mRNA therapeutics for global needs”.
Arnav – which has RNA right there in the name – in Sanskrit means ‘ocean.’ An ocean has no discernible borders, and Tiwari is working to build a biotech company that eliminates borders in equitable access to mRNA-based therapeutics and vaccines.
With this mission in mind, Arnav is developing mRNA-based, broad-spectrum antivirals as well as vaccines against pandemic potential viruses before the next pandemic hits. Arnav has recently entered in a collaboration with Sartorius BIA Separations, a company based on Slovenia, to advance their mRNA pipeline. While building its own mRNA therapeutics pipeline, Arnav is also helping other scientists explore mRNA as an alternative therapeutic and vaccine platform through its contract services.
“I think of the vaccine scientist who makes his medicine using proteins, but would like to explore the mRNA option,” Tiwari posits. “Maybe he doesn’t want to make the full jump into it. That’s where we come in, helping to drive interest in this field and help that scientist compare his traditional vaccines to see what mRNA vaccines looks like.”
She has all the equipment and instruments that she needs at BioSpark Labs and was one of the first start-ups to put down roots there. So far, it’s been the perfect partnership, Tiwari said, adding, “It kind of feels like BioSpark and Arnav are growing up together.”
News Contact
Writer: Jerry Grillo
Feb. 09, 2023
Coral reef conservation is a steppingstone to protect marine biodiversity and life in the ocean as we know it. The health of coral also has huge societal implications: reef ecosystems provide sustenance and livelihoods for millions of people around the world. Conserving biodiversity in reef areas is both a social issue and a marine biodiversity priority.
In the face of climate change, Annalisa Bracco, professor in the School of Earth and Atmospheric Sciences at Georgia Institute of Technology, and Lyuba Novi, a postdoctoral researcher, offer a new methodology that could revolutionize how conservationists monitor coral. The researchers applied machine learning tools to study how climate impacts connectivity and biodiversity in the Pacific Ocean’s Coral Triangle — the most diverse and biologically complex marine ecosystem on the planet. Their research, recently published in Nature Communications Biology, overcomes time and resource barriers to contextualize the biodiversity of the Coral Triangle, while offering hope for better monitoring and protection in the future.
“We saw that the biodiversity of the Coral Triangle is incredibly dynamic,” Bracco said. “For a long time, it has been postulated that this is due to sea level change and distribution of land masses, but we are now starting to understand that there is more to the story.”
Connectivity refers to the conditions that allow different ecosystems to exchange genetic material such as eggs, larvae, or the young. Ocean currents spread genetic material and also create the dynamics that allow a body of water — and thus ecosystems — to maintain consistent chemical, biological, and physical properties. If coral larvae are spread to an ecoregion where the conditions are very similar to the original location, the larvae can start a new coral.
Bracco wanted to see how climate, and specifically the El Niño Southern Oscillation (ENSO) in its phases — El Niño, La Niña, and neutral conditions — impacts connectivity in the Coral Triangle. Climate events that move large masses of warm water in the Pacific Ocean bring enormous changes and have been known to exacerbate coral bleaching, in which corals turn white due to environmental stressors and become vulnerable to disease.
“Biologists collect data in situ, which is extremely important,” Bracco said. “But it’s not possible to monitor enormous regions in situ for many years — that would require a constant presence of scuba divers. So, figuring out how different ocean regions and large marine ecosystems are connected over time, especially in terms of foundational species, becomes important.”
Machine Learning for Discovering Connectivity
Years ago, Bracco and collaborators developed a tool, Delta Maps, that uses machine learning to identify “domains,” or regions within any kind of system that share the same dynamic. Bracco initially used it to analyze domains of climate variability in models but also suspected it could be used to study ecoregions in the ocean.
For this study, they used the tool to map out domains of connectivity in the Coral Triangle using 30 years of sea surface temperature data. Sea surface temperatures change in response to ocean currents over scales of weeks and months and across distances of tens of kilometers. These changes are relevant to coral connectivity, so the researchers built their machine learning tool based on this observation, using changes in surface ocean temperature to identify regions connected by currents. They also separated the time periods that they were considering into three categories: El Niño events, La Niña events, and neutral or “normal” times, painting a picture of how connectivity was impacted during major climate events in particular ecoregions.
Novi then applied a ranking system to the different ecoregions they identified. She used rank page centrality, a machine learning tool that was invented to rank webpages on the internet, on top of Delta Maps to identify which coral ecoregions were most strongly connected and able to receive the most coral larvae from other regions. Those regions would be the ones most likely sustain and survive through a bleaching event.
Climate Dynamics and Biodiversity
Bracco and Novi found that climate dynamics have contributed to biodiversity because of the way climate introduces variability to the currents in the equatorial Pacific Ocean. The researchers realized that alternation of El Niño and La Niña events has allowed for enormous genetic exchanges between the Indian and Pacific Oceans and enabled the ecosystems to survive through a variety of different climate situations.
“There is never an identical connection between ecoregions in all ENSO phases,” Bracco said. “In other parts of the world ocean, coral reefs are connected through a fixed, often small, number of ecoregions, and if you eliminate this fixed number of connections by bleaching all connected reefs, you will not be able to rebuild the corals in any of them. But in the Pacific the connections are changing all the time and are so dynamic that soon enough the bleached reef will receive larvae from completely different ecoregions in a different ENSO phase.”
They also concluded that, because of the Coral Triangle’s dynamic climate component, there is more possibility for rebuilding biodiversity there than anywhere else on the planet. And that the evolution of biodiversity in the Coral Triangle is not only linked to landmasses or sea levels but also to the evolution of ENSO through geological times. The researchers found that though ENSO causes coral bleaching, it has helped the Coral Triangle become so rich in biodiversity.
Better Monitoring Opportunities
Because coral reef survival has been designated a priority by the United Nations Sustainable Development Goals, Bracco and Novi’s research is poised to have broad applications. The researchers’ method identified which ecoregions conservationists should try hardest to protect and also the regions that conservationists could expect to have the most luck with protection measures. Their methodology can also help to identify which regions should be monitored more and the ones that could be considered lower priority for now due to the ways they are currently thriving.
“This research opens a lot of possibilities for better monitoring strategies, and especially how to monitor given a limited amount of resources and money,” Bracco said. “As of now, coral monitoring often happens when groups have a limited amount of funding to apply to a very specific localized region. We hope our method can be used to create a better monitoring over larger scales of time and space.”
CITATION: Novi, L., Bracco, A. “Machine learning prediction of connectivity, biodiversity and resilience in the Coral Triangle.” Commun Biol 5, 1359 (2022).
News Contact
Catherine Barzler, Senior Research Writer/Editor
Pagination
- Previous page
- 2 Page 2
- Next page