Apr. 25, 2025
A Georgia Tech doctoral student’s dissertation could help physicians diagnose neuropsychiatric disorders, including schizophrenia, autism, and Alzheimer’s disease. The new approach leverages data science and algorithms instead of relying on traditional methods like cognitive tests and image scans.
Ph.D. candidate Md Abdur Rahaman’s dissertation studies brain data to understand how changes in brain activity shape behavior.
Computational tools Rahaman developed for his dissertation look for informative patterns between the brain and behavior. Successful tests of his algorithms show promise to help doctors diagnose mental health disorders and design individualized treatment plans for patients.
“I've always been fascinated by the human brain and how it defines who we are,” Rahaman said.
“The fact that so many people silently suffer from neuropsychiatric disorders, while our understanding of the brain remains limited, inspired me to develop tools that bring greater clarity to this complexity and offer hope through more compassionate, data-driven care.”
Rahaman’s dissertation introduces a framework focusing on granular factoring. This computing technique stratifies brain data into smaller, localized subgroups, making it easier for computers and researchers to study data and find meaningful patterns.
Granular factoring overcomes the challenges of size and heterogeneity in neurological data science. Brain data is obtained from neuroimaging, genomics, behavioral datasets, and other sources. The large size of each source makes it a challenge to study them individually, let alone analyze them simultaneously, to find hidden inferences.
Rahaman’s research allows researchers and physicians to move past one-size-fits-all approaches. Instead of manually reviewing tests and scans, algorithms look for patterns and biomarkers in the subgroups that otherwise go undetected, especially ones that indicate neuropsychiatric disorders.
“My dissertation advances the frontiers of computational neuroscience by introducing scalable and interpretable models that navigate brain heterogeneity to reveal how neural dynamics shape behavior,” Rahaman said.
“By uncovering subgroup-specific patterns, this work opens new directions for understanding brain function and enables more precise, personalized approaches to mental health care.”
Rahaman defended his dissertation on April 14, the final step in completing his Ph.D. in computational science and engineering. He will graduate on May 1 at Georgia Tech’s Ph.D. Commencement.
After walking across the stage at McCamish Pavilion, Rahaman’s next step in his career is to go to Amazon, where he will work in the generative artificial intelligence (AI) field.
Graduating from Georgia Tech is the summit of an educational trek spanning over a decade. Rahaman hails from Bangladesh where he graduated from Chittagong University of Engineering and Technology in 2013. He attained his master’s from the University of New Mexico in 2019 before starting at Georgia Tech.
“Munna is an amazingly creative researcher,” said Vince Calhoun, Rahman’s advisor. Calhoun is the founding director of the Translational Research in Neuroimaging and Data Science Center (TReNDS).
TReNDS is a tri-institutional center spanning Georgia Tech, Georgia State University, and Emory University that develops analytic approaches and neuroinformatic tools. The center aims to translate the approaches into biomarkers that address areas of brain health and disease.
“His work is moving the needle in our ability to leverage multiple sources of complex biological data to improve understanding of neuropsychiatric disorders that have a huge impact on an individual’s livelihood,” said Calhoun.
News Contact
Bryant Wine, Communications Officer
bryant.wine@cc.gatech.edu
Apr. 23, 2025
Georgia Tech professors Michelle LaPlaca and W. Hong Yeo have been selected as recipients of Peterson Professorships with the Children’s Healthcare of Atlanta Pediatric Technology Center (PTC) at Georgia Tech. The professorships, supported by the G.P. “Bud” Peterson and Valerie H. Peterson Faculty Endowment Fund, are meant to further energize the Georgia Tech and Children’s partnership by engaging and empowering researchers involved in pediatrics.
In a joint statement, PTC co-directors Wilbur Lam and Stanislav Emelianov said, “The appointment of Dr. LaPlaca and Dr. Yeo as Peterson Professors exemplifies the vision of Bud and Valerie Peterson — advancing innovation and collaboration through the Pediatric Technology Center to bring breakthrough ideas from the lab to the bedside, improving the lives of children and transforming healthcare.”
LaPlaca is a professor and associate chair for Faculty Development in the Department of Biomedical Engineering, a joint department between Georgia Tech and Emory University. Her research is focused on traumatic brain injury and concussion, concentrating on sources of heterogeneity and clinical translation. Specifically, she is working on biomarker discovery, the role of the glymphatic system, and novel virtual reality neurological assessments.
“I am thrilled to be chosen as one of the Peterson Professors and appreciate Bud and Valerie Peterson’s dedication to pediatric research,” she said. “The professorship will allow me to broaden research in pediatric concussion assessment and college student concussion awareness, as well as to identify biomarkers in experimental models of brain injury.”
In addition to the research lab, LaPlaca will work with an undergraduate research class called Concussion Connect, which is part of the Vertically Integrated Projects program at Georgia Tech.
“Through the PTC, Georgia Tech and Children’s will positively impact brain health in Georgia’s pediatric population,” said LaPlaca.
Yeo is the Harris Saunders, Jr. Professor in the George W. Woodruff School of Mechanical Engineering and the director of the Wearable Intelligent Systems and Healthcare Center at Georgia Tech. His research focuses on nanomanufacturing and membrane electronics to develop soft biomedical devices aimed at improving disease diagnostics, therapeutics, and rehabilitation.
“I am truly honored to be awarded the Peterson Professorship from the Children’s PTC at Georgia Tech,” he said. “This recognition will greatly enhance my research efforts in developing soft bioelectronics aimed at advancing pediatric healthcare, as well as expand education opportunities for the next generation of undergraduate and graduate students interested in creating innovative medical devices that align seamlessly with the recent NSF Research Traineeship grant I received. I am eager to contribute to the dynamic partnership between Georgia Tech and Children’s Healthcare of Atlanta and to empower innovative solutions that will improve the lives of children.”
The Peterson Professorships honor the former Georgia Tech President and First Lady, whose vision for the importance of research in improving pediatric healthcare has had an enormous positive impact on the care of pediatric patients in our state and region.
The Children’s PTC at Georgia Tech brings clinical experts from Children’s together with Georgia Tech scientists and engineers to develop technological solutions to problems in the health and care of children. Children’s PTC provides extraordinary opportunities for interdisciplinary collaboration in pediatrics, creating breakthrough discoveries that often can only be found at the intersection of multiple disciplines. These collaborations also allow us to bring discoveries to the clinic and the bedside, thereby enhancing the lives of children and young adults. The mission of the PTC is to establish the world’s leading program in the development of technological solutions for children’s health, focused on three strategic areas that will have a lasting impact on Georgia’s kids and beyond.
Apr. 10, 2025
In a groundbreaking study published in Nature, researchers from Georgia Tech and Emory University have developed a new model that could enable precise, life-saving medication delivery for blood clot patients. The novel technique uses a 3D microchip
Wilbur Lam, professor at Georgia Tech and Emory University, and a clinician at Children’s Healthcare of Atlanta, led the study. He worked closely with Yongzhi Qiu, an assistant professor in the Department of Pediatrics at Emory University School of Medicine.
The significance of the thromboinflammation-on-a-chip model, is that it mimics clots in a human-like way, allowing them to last for months and resolve naturally. This model helps track blood clots and more effectively test treatments for conditions including sickle cell anemia, strokes, and heart attacks.
News Contact
Amelia Neumeister | Research Communications Program Manager
Dec. 18, 2024
As we go through our daily routines of work, chores, errands and leisure pursuits, most of us take our mobility for granted. Conversely, many people suffer from permanent or temporary mobility issues due to neurological disorders, stroke, injury, and age-related causes. Research in the field of robotic exoskeletons has shown significant potential to provide assistive support for patients with permanent mobility constraints, as well as an effective additional tool for rehabilitation and recovery after injury.
Though the field has made great progress in the hardware and devices for these assistive technologies, there are limitations in ease of use and in the ability to move from walking to running, from flat ground to slopes and stairs, and across different terrains. Recent developments to create exoskeleton controllers that are more responsive to the user’s environment via user-based variables such as gait and slope calculations provide rapid yet imprecise outputs. More recent inquiry into data-driven improvements such as vision-based labeling and classification are extremely promising additions in the goal to develop a true synchronous user and device interface. A major hindrance to this data-driven approach is the need for burdensome mounted cameras and on-board computing to allow for real-time in use adjustments to the environmental terrain encountered.
In order to address these barriers, Aaron Young, Associate Professor in the Woodruff School of Mechanical Engineering and Director of the Exoskeleton and Prosthetic Intelligent Controls (EPIC) Lab, and Dawit Lee, Postdoctoral Scholar at Stanford, have created an artificial intelligence (AI)-based universal exoskeleton controller that uses information from onboard mechanical sensors without the added weight and complexity of mounted vision based systems. The new work, published in Science Advances (Link to Be Added), presents a controller that holistically captures the major variations encountered during community walking in real-time. The team combined data from the Americans with Disabilities Act (ADA) building guidelines that characterize ambulatory terrains in slope level degrees with a gait phase estimator to achieve dynamic switching of assistance types between multiple terrains and slopes and delivery to the user with little to no delay.
In this work, we have created a new, open-source knee exoskeleton design that is intended to support community mobility. Knee assist devices have tremendous value in activities such as sit-to-stand, stairs, and ramps where we use our biological knees substantially to accomplish these tasks. The neat accomplishment in this work is that by leveraging AI, we avoid the need to classify these different modes discretely but rather have a single continuous variable (in this case rise over run of the surface) to enable continuous and unified control over common ambulatory tasks such as walking, stairs, and ramps. We demonstrate that on novel users of the device, we can track both the environment and the user’s gait state with very high accuracy out of the lab in community settings. It is an exciting time in the field as we see more studies, such as this one, showing promise in tackling real-world mobility challenges
The assistance approach using our intelligent controller, presented in this work, provides users with support at the right timing and with a magnitude that closely matches the varying biomechanical effort they produce as they move through the community. Our assistance approach was preferred for community navigation and was more effective in reducing the user’s energy consumption compared to conventional methods. We also open-sourced the design of the robotic knee exoskeleton hardware and the dataset used to train the models with this publication which allows other researchers to build upon our developments and further advance the field. This work demonstrates an exciting example of AI integration into a wearable robotic system, showcasing its successful outcomes and significant potential.
- Dawit Lee; Postdoctoral Scholar, Stanford
Using this combination of a universal slope estimator and a gait phase estimator, the team achieved results in the dynamic modulation of exoskeleton assistance that have never been achieved by previous approaches and moves the field closer to creating an adaptive and effective assistive technology that seamlessly integrates into the daily lives of individuals, promoting enhanced mobility and overall well-being. This work also has the potential to enable a mode-specific assistance approach tailored to the user’s specific biomechanical needs.
- Christa M. Ernst; Research Communications Program Manager
Original Publication
Dawit Lee, Sanghyub Lee, and Aaron J. Young, “AI-Driven Universal Lower-Limb Exoskeleton System for Community Ambulation,” Science Advances
Prior Related Work
D. Lee, I. Kang, D. D. Molinaro, A. Yu, A. J. Young, Real-time user-independent slope prediction using deep learning for modulation of robotic knee exoskeleton assistance. IEEE Robot. Autom. Lett. 6, 3995–4000 (2021).
Funding Provided by
NIH Director’s New Innovator Award DP2-HD111709
Dec. 04, 2024
A new machine learning (ML) model from Georgia Tech could protect communities from diseases, better manage electricity consumption in cities, and promote business growth, all at the same time.
Researchers from the School of Computational Science and Engineering (CSE) created the Large Pre-Trained Time-Series Model (LPTM) framework. LPTM is a single foundational model that completes forecasting tasks across a broad range of domains.
Along with performing as well or better than models purpose-built for their applications, LPTM requires 40% less data and 50% less training time than current baselines. In some cases, LPTM can be deployed without any training data.
The key to LPTM is that it is pre-trained on datasets from different industries like healthcare, transportation, and energy. The Georgia Tech group created an adaptive segmentation module to make effective use of these vastly different datasets.
The Georgia Tech researchers will present LPTM in Vancouver, British Columbia, Canada, at the 2024 Conference on Neural Information Processing Systems (NeurIPS 2024). NeurIPS is one of the world’s most prestigious conferences on artificial intelligence (AI) and ML research.
“The foundational model paradigm started with text and image, but people haven’t explored time-series tasks yet because those were considered too diverse across domains,” said B. Aditya Prakash, one of LPTM’s developers.
“Our work is a pioneer in this new area of exploration where only few attempts have been made so far.”
[MICROSITE: Georgia Tech at NeurIPS 2024]
Foundational models are trained with data from different fields, making them powerful tools when assigned tasks. Foundational models drive GPT, DALL-E, and other popular generative AI platforms used today. LPTM is different though because it is geared toward time-series, not text and image generation.
The Georgia Tech researchers trained LPTM on data ranging from epidemics, macroeconomics, power consumption, traffic and transportation, stock markets, and human motion and behavioral datasets.
After training, the group pitted LPTM against 17 other models to make forecasts as close to nine real-case benchmarks. LPTM performed the best on five datasets and placed second on the other four.
The nine benchmarks contained data from real-world collections. These included the spread of influenza in the U.S. and Japan, electricity, traffic, and taxi demand in New York, and financial markets.
The competitor models were purpose-built for their fields. While each model performed well on one or two benchmarks closest to its designed purpose, the models ranked in the middle or bottom on others.
In another experiment, the Georgia Tech group tested LPTM against seven baseline models on the same nine benchmarks in zero-shot forecasting tasks. Zero-shot means the model is used out of the box and not given any specific guidance during training. LPTM outperformed every model across all benchmarks in this trial.
LPTM performed consistently as a top-runner on all nine benchmarks, demonstrating the model’s potential to achieve superior forecasting results across multiple applications with less and resources.
“Our model also goes beyond forecasting and helps accomplish other tasks,” said Prakash, an associate professor in the School of CSE.
“Classification is a useful time-series task that allows us to understand the nature of the time-series and label whether that time-series is something we understand or is new.”
One reason traditional models are custom-built to their purpose is that fields differ in reporting frequency and trends.
For example, epidemic data is often reported weekly and goes through seasonal peaks with occasional outbreaks. Economic data is captured quarterly and typically remains consistent and monotone over time.
LPTM’s adaptive segmentation module allows it to overcome these timing differences across datasets. When LPTM receives a dataset, the module breaks data into segments of different sizes. Then, it scores all possible ways to segment data and chooses the easiest segment from which to learn useful patterns.
LPTM’s performance, enhanced through the innovation of adaptive segmentation, earned the model acceptance to NeurIPS 2024 for presentation. NeurIPS is one of three primary international conferences on high-impact research in AI and ML. NeurIPS 2024 occurs Dec. 10-15.
Ph.D. student Harshavardhan Kamarthi partnered with Prakash, his advisor, on LPTM. The duo are among the 162 Georgia Tech researchers presenting over 80 papers at the conference.
Prakash is one of 46 Georgia Tech faculty with research accepted at NeurIPS 2024. Nine School of CSE faculty members, nearly one-third of the body, are authors or co-authors of 17 papers accepted at the conference.
Along with sharing their research at NeurIPS 2024, Prakash and Kamarthi released an open-source library of foundational time-series modules that data scientists can use in their applications.
“Given the interest in AI from all walks of life, including business, social, and research and development sectors, a lot of work has been done and thousands of strong papers are submitted to the main AI conferences,” Prakash said.
“Acceptance of our paper speaks to the quality of the work and its potential to advance foundational methodology, and we hope to share that with a larger audience.”
News Contact
Bryant Wine, Communications Officer
bryant.wine@cc.gatech.edu
Nov. 13, 2024
Amino acids are essential for nearly every process in the human body. Often referred to as ‘the building blocks of life,’ they are also critical for commercial use in products ranging from pharmaceuticals and dietary supplements, to cosmetics, animal feed, and industrial chemicals.
And while our bodies naturally make amino acids, manufacturing them for commercial use can be costly — and that process often emits greenhouse gasses like carbon dioxide (CO2).
In a landmark study, a team of researchers has created a first-of-its kind methodology for synthesizing amino acids that uses more carbon than it emits. The research also makes strides toward making the system cost-effective and scalable for commercial use.
“To our knowledge, it’s the first time anyone has synthesized amino acids in a carbon-negative way using this type of biocatalyst,” says lead corresponding author Pamela Peralta-Yahya, who emphasizes that the system provides a win-win for industry and environment. “Carbon dioxide is readily available, so it is a low-cost feedstock — and the system has the added bonus of removing a powerful greenhouse gas from the atmosphere, making the synthesis of amino acids environmentally friendly, too.”
The study, “Carbon Negative Synthesis of Amino Acids Using a Cell-Free-Based Biocatalyst,” published today in ACS Synthetic Biology, is publicly available. The research was led by Georgia Tech in collaboration with the University of Washington, Pacific Northwest National Laboratory, and the University of Minnesota.
The Georgia Tech research contingent includes Peralta-Yahya, a professor with joint appointments in the School of Chemistry and Biochemistry and School of Chemical and Biomolecular Engineering (ChBE); first author Shaafique Chowdhury, a Ph.D. student in ChBE; Ray Westenberg, a Ph.D student in Bioengineering; and Georgia Tech alum Kimberly Wennerholm (B.S. ChBE ’23).
Costly chemicals
There are two key challenges to synthesizing amino acids on a large scale: the cost of materials, and the speed at which the system can generate amino acids.
While many living systems like cyanobacteria can synthesize amino acids from CO2, the rate at which they do it is too slow to be harnessed for industrial applications, and these systems can only synthesize a limited number of chemicals.
Currently, most commercial amino acids are made using bioengineered microbes. “These specially designed organisms convert sugar or plant biomass into fuel and chemicals,” explains first author Chowdhury, “but valuable food resources are consumed if sugar is used as the feedstock — and pre-processing plant biomass is costly.” These processes also release CO2 as a byproduct.
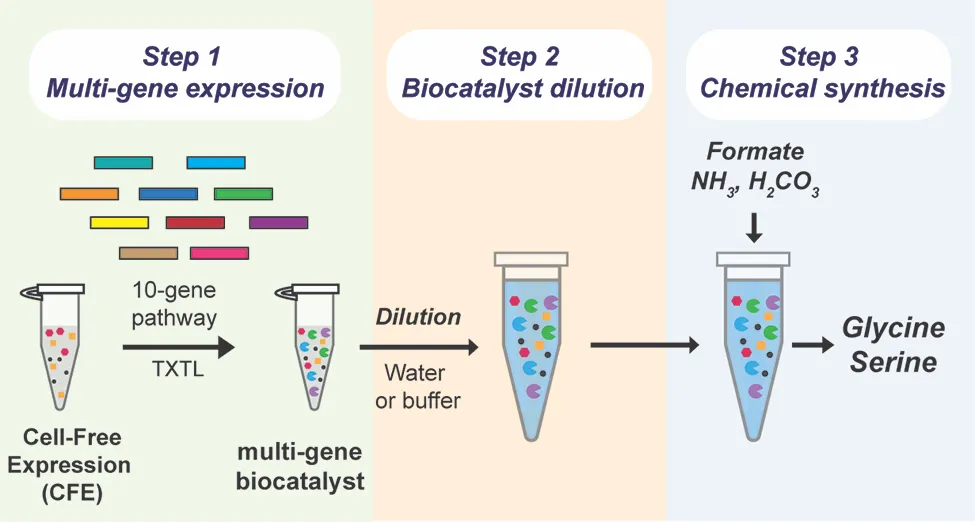
Chowdhury says the team was curious “if we could develop a commercially viable system that could use carbon dioxide as a feedstock. We wanted to build a system that could quickly and efficiently convert CO2 into critical amino acids, like glycine and serine.”
The team was particularly interested in what could be accomplished by a ‘cell-free’ system that leveraged some process of a cellular system — but didn’t actually involve living cells, Peralta-Yahya says, adding that systems using living cells need to use part of their CO2 to fuel their own metabolic processes, including cell growth, and have not yet produced sufficient quantities of amino acids.
“Part of what makes a cell-free system so efficient,” Westenberg explains, “is that it can use cellular enzymes without needing the cells themselves. By generating the enzymes and combining them in the lab, the system can directly convert carbon dioxide into the desired chemicals. Because there are no cells involved, it doesn’t need to use the carbon to support cell growth — which vastly increases the amount of amino acids the system can produce.”
A novel solution
While scientists have used cell-free systems before, one of the necessary chemicals, the cell lysate biocatalyst, is extremely costly. For a cell-free system to be economically viable at scale, the team needed to limit the amount of cell lysate the system needed.
After creating the ten enzymes necessary for the reaction, the team attempted to dilute the biocatalyst using a technique called ‘volumetric expansion.’ “We found that the biocatalyst we used was active even after being diluted 200-fold,” Peralta-Yahya explains. “This allows us to use significantly less of this high-cost material — while simultaneously increasing feedstock loading and amino acid output.”
It’s a novel application of a cell-free system, and one with the potential to transform both how amino acids are produced, and the industry’s impact on our changing climate.
“This research provides a pathway for making this method cost-effective and scalable,” Peralta-Yahya says. “This system might one day be used to make chemicals ranging from aromatics and terpenes, to alcohols and polymers, and all in a way that not only reduces our carbon footprint, but improves it.”
Funding: Advanced Research Project Agency-Energy (ARPA-E), U.S. Department of Energy and the U.S. Department of Energy, Office of Science, Biological and Environmental Research Program.
News Contact
Written by Selena Langner
Oct. 22, 2024
For his work creating new kinds of drug delivery techniques and bringing those technologies to patients, Mark Prausnitz is one of the new members of the National Academy of Medicine (NAM).
The Academy announced his election Oct. 21 alongside 99 others. Membership in NAM is considered one of the highest recognitions in health and medicine, reserved for those who’ve made major contributions to healthcare, medical sciences, and public health. The roster is small: only 2,400 or so individuals have been honored.
“It’s an honor to be elected to the National Academy of Medicine and have the work of our team at Georgia Tech recognized in this way,” said Prausnitz, Regents’ Professor and J. Erskine Love Jr. Chair in the School of Chemical and Biomolecular Engineering.
The Academy cited Prausnitz for innovating microneedle and other advanced drug delivery technologies. He also was honored for translating those methods and devices into clinical trials and products and founding companies to bring the advances to patients. NAM praised Prausnitz for “inspiring students to be creative and impactful engineers.”
News Contact
Joshua Stewart
College of Engineering
Oct. 16, 2024
A new surgery planning tool powered by augmented reality (AR) is in development for doctors who need closer collaboration when planning heart operations. Promising results from a recent usability test have moved the platform one step closer to everyday use in hospitals worldwide.
Georgia Tech researchers partnered with medical experts from Children’s Healthcare of Atlanta (CHOA) to develop and test ARCollab. The iOS-based app leverages advanced AR technologies to let doctors collaborate together and interact with a patient’s 3D heart model when planning surgeries.
The usability evaluation demonstrates the app’s effectiveness, finding that ARCollab is easy to use and understand, fosters collaboration, and improves surgical planning.
“This tool is a step toward easier collaborative surgical planning. ARCollab could reduce the reliance on physical heart models, saving hours and even days of time while maintaining the collaborative nature of surgical planning,” said M.S. student Pratham Mehta, the app’s lead researcher.
“Not only can it benefit doctors when planning for surgery, it may also serve as a teaching tool to explain heart deformities and problems to patients.”
Two cardiologists and three cardiothoracic surgeons from CHOA tested ARCollab. The two-day study ended with the doctors taking a 14-question survey assessing the app’s usability. The survey also solicited general feedback and top features.
The Georgia Tech group determined from the open-ended feedback that:
- ARCollab enables new collaboration capabilities that are easy to use and facilitate surgical planning.
- Anchoring the model to a physical space is important for better interaction.
- Portability and real-time interaction are crucial for collaborative surgical planning.
Users rated each of the 14 questions on a 7-point Likert scale, with one being “strongly disagree” and seven being “strongly agree.” The 14 questions were organized into five categories: overall, multi-user, model viewing, model slicing, and saving and loading models.
The multi-user category attained the highest rating with an average of 6.65. This included a unanimous 7.0 rating that it was easy to identify who was controlling the heart model in ARCollab. The scores also showed it was easy for users to connect with devices, switch between viewing and slicing, and view other users’ interactions.
The model slicing category received the lowest, but formidable, average of 5.5. These questions assessed ease of use and understanding of finger gestures and usefulness to toggle slice direction.
Based on feedback, the researchers will explore adding support for remote collaboration. This would assist doctors in collaborating when not in a shared physical space. Another improvement is extending the save feature to support multiple states.
“The surgeons and cardiologists found it extremely beneficial for multiple people to be able to view the model and collaboratively interact with it in real-time,” Mehta said.
The user study took place in a CHOA classroom. CHOA also provided a 3D heart model for the test using anonymous medical imaging data. Georgia Tech’s Institutional Review Board (IRB) approved the study and the group collected data in accordance with Institute policies.
The five test participants regularly perform cardiovascular surgical procedures and are employed by CHOA.
The Georgia Tech group provided each participant with an iPad Pro with the latest iOS version and the ARCollab app installed. Using commercial devices and software meets the group’s intentions to make the tool universally available and deployable.
“We plan to continue iterating ARCollab based on the feedback from the users,” Mehta said.
“The participants suggested the addition of a ‘distance collaboration’ mode, enabling doctors to collaborate even if they are not in the same physical environment. This allows them to facilitate surgical planning sessions from home or otherwise.”
The Georgia Tech researchers are presenting ARCollab and the user study results at IEEE VIS 2024, the Institute of Electrical and Electronics Engineers (IEEE) visualization conference.
IEEE VIS is the world’s most prestigious conference for visualization research and the second-highest rated conference for computer graphics. It takes place virtually Oct. 13-18, moved from its venue in St. Pete Beach, Florida, due to Hurricane Milton.
The ARCollab research group's presentation at IEEE VIS comes months after they shared their work at the Conference on Human Factors in Computing Systems (CHI 2024).
Undergraduate student Rahul Narayanan and alumni Harsha Karanth (M.S. CS 2024) and Haoyang (Alex) Yang (CS 2022, M.S. CS 2023) co-authored the paper with Mehta. They study under Polo Chau, a professor in the School of Computational Science and Engineering.
The Georgia Tech group partnered with Dr. Timothy Slesnick and Dr. Fawwaz Shaw from CHOA on ARCollab’s development and user testing.
"I'm grateful for these opportunities since I get to showcase the team's hard work," Mehta said.
“I can meet other like-minded researchers and students who share these interests in visualization and human-computer interaction. There is no better form of learning.”
News Contact
Bryant Wine, Communications Officer
bryant.wine@cc.gatech.edu
Jul. 15, 2024
Hepatic, or liver, disease affects more than 100 million people in the U.S. About 4.5 million adults (1.8%) have been diagnosed with liver disease, but it is estimated that between 80 and 100 million adults in the U.S. have undiagnosed fatty liver disease in varying stages. Over time, undiagnosed and untreated hepatic diseases can lead to cirrhosis, a severe scarring of the liver that cannot be reversed.
Most hepatic diseases are chronic conditions that will be present over the life of the patient, but early detection improves overall health and the ability to manage specific conditions over time. Additionally, assessing patients over time allows for effective treatments to be adjusted as necessary. The standard protocol for diagnosis, as well as follow-up tissue assessment, is a biopsy after the return of an abnormal blood test, but biopsies are time-consuming and pose risks for the patient. Several non-invasive imaging techniques have been developed to assess the stiffness of liver tissue, an indication of scarring, including magnetic resonance elastography (MRE).
MRE combines elements of ultrasound and MRI imaging to create a visual map showing gradients of stiffness throughout the liver and is increasingly used to diagnose hepatic issues. MRE exams, however, can fail for many reasons, including patient motion, patient physiology, imaging issues, and mechanical issues such as improper wave generation or propagation in the liver. Determining the success of MRE exams depends on visual inspection of technologists and radiologists. With increasing work demands and workforce shortages, providing an accurate, automated way to classify image quality will create a streamlined approach and reduce the need for repeat scans.
Professor Jun Ueda in the George W. Woodruff School of Mechanical Engineering and robotics Ph.D. student Heriberto Nieves, working with a team from the Icahn School of Medicine at Mount Sinai, have successfully applied deep learning techniques for accurate, automated quality control image assessment. The research, “Deep Learning-Enabled Automated Quality Control for Liver MR Elastography: Initial Results,” was published in the Journal of Magnetic Resonance Imaging.
Using five deep learning training models, an accuracy of 92% was achieved by the best-performing ensemble on retrospective MRE images of patients with varied liver stiffnesses. The team also achieved a return of the analyzed data within seconds. The rapidity of image quality return allows the technician to focus on adjusting hardware or patient orientation for re-scan in a single session, rather than requiring patients to return for costly and timely re-scans due to low-quality initial images.
This new research is a step toward streamlining the review pipeline for MRE using deep learning techniques, which have remained unexplored compared to other medical imaging modalities. The research also provides a helpful baseline for future avenues of inquiry, such as assessing the health of the spleen or kidneys. It may also be applied to automation for image quality control for monitoring non-hepatic conditions, such as breast cancer or muscular dystrophy, in which tissue stiffness is an indicator of initial health and disease progression. Ueda, Nieves, and their team hope to test these models on Siemens Healthineers magnetic resonance scanners within the next year.
Publication
Nieves-Vazquez, H.A., Ozkaya, E., Meinhold, W., Geahchan, A., Bane, O., Ueda, J. and Taouli, B. (2024), Deep Learning-Enabled Automated Quality Control for Liver MR Elastography: Initial Results. J Magn Reson Imaging. https://doi.org/10.1002/jmri.29490
Prior Work
Robotically Precise Diagnostics and Therapeutics for Degenerative Disc Disorder
Related Material
Editorial for “Deep Learning-Enabled Automated Quality Control for Liver MR Elastography: Initial Results”
News Contact
Christa M. Ernst |
Research Communications Program Manager |
Topic Expertise: Robotics, Data Sciences, Semiconductor Design & Fab |
May. 15, 2024
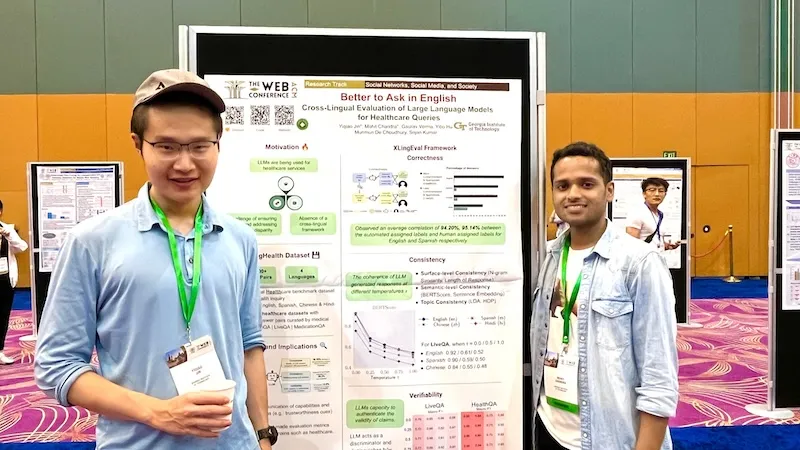
Georgia Tech researchers say non-English speakers shouldn’t rely on chatbots like ChatGPT to provide valuable healthcare advice.
A team of researchers from the College of Computing at Georgia Tech has developed a framework for assessing the capabilities of large language models (LLMs).
Ph.D. students Mohit Chandra and Yiqiao (Ahren) Jin are the co-lead authors of the paper Better to Ask in English: Cross-Lingual Evaluation of Large Language Models for Healthcare Queries.
Their paper’s findings reveal a gap between LLMs and their ability to answer health-related questions. Chandra and Jin point out the limitations of LLMs for users and developers but also highlight their potential.
Their XLingEval framework cautions non-English speakers from using chatbots as alternatives to doctors for advice. However, models can improve by deepening the data pool with multilingual source material such as their proposed XLingHealth benchmark.
“For users, our research supports what ChatGPT’s website already states: chatbots make a lot of mistakes, so we should not rely on them for critical decision-making or for information that requires high accuracy,” Jin said.
“Since we observed this language disparity in their performance, LLM developers should focus on improving accuracy, correctness, consistency, and reliability in other languages,” Jin said.
Using XLingEval, the researchers found chatbots are less accurate in Spanish, Chinese, and Hindi compared to English. By focusing on correctness, consistency, and verifiability, they discovered:
- Correctness decreased by 18% when the same questions were asked in Spanish, Chinese, and Hindi.
- Answers in non-English were 29% less consistent than their English counterparts.
- Non-English responses were 13% overall less verifiable.
XLingHealth contains question-answer pairs that chatbots can reference, which the group hopes will spark improvement within LLMs.
The HealthQA dataset uses specialized healthcare articles from the popular healthcare website Patient. It includes 1,134 health-related question-answer pairs as excerpts from original articles.
LiveQA is a second dataset containing 246 question-answer pairs constructed from frequently asked questions (FAQs) platforms associated with the U.S. National Institutes of Health (NIH).
For drug-related questions, the group built a MedicationQA component. This dataset contains 690 questions extracted from anonymous consumer queries submitted to MedlinePlus. The answers are sourced from medical references, such as MedlinePlus and DailyMed.
In their tests, the researchers asked over 2,000 medical-related questions to ChatGPT-3.5 and MedAlpaca. MedAlpaca is a healthcare question-answer chatbot trained in medical literature. Yet, more than 67% of its responses to non-English questions were irrelevant or contradictory.
“We see far worse performance in the case of MedAlpaca than ChatGPT,” Chandra said.
“The majority of the data for MedAlpaca is in English, so it struggled to answer queries in non-English languages. GPT also struggled, but it performed much better than MedAlpaca because it had some sort of training data in other languages.”
Ph.D. student Gaurav Verma and postdoctoral researcher Yibo Hu co-authored the paper.
Jin and Verma study under Srijan Kumar, an assistant professor in the School of Computational Science and Engineering, and Hu is a postdoc in Kumar’s lab. Chandra is advised by Munmun De Choudhury, an associate professor in the School of Interactive Computing.
The team will present their paper at The Web Conference, occurring May 13-17 in Singapore. The annual conference focuses on the future direction of the internet. The group’s presentation is a complimentary match, considering the conference's location.
English and Chinese are the most common languages in Singapore. The group tested Spanish, Chinese, and Hindi because they are the world’s most spoken languages after English. Personal curiosity and background played a part in inspiring the study.
“ChatGPT was very popular when it launched in 2022, especially for us computer science students who are always exploring new technology,” said Jin. “Non-native English speakers, like Mohit and I, noticed early on that chatbots underperformed in our native languages.”
School of Interactive Computing communications officer Nathan Deen and School of Computational Science and Engineering communications officer Bryant Wine contributed to this report.
News Contact
Bryant Wine, Communications Officer
bryant.wine@cc.gatech.edu
Nathan Deen, Communications Officer
ndeen6@cc.gatech.edu
Pagination
- 1 Page 1
- Next page