Feb. 09, 2023
Coral reef conservation is a steppingstone to protect marine biodiversity and life in the ocean as we know it. The health of coral also has huge societal implications: reef ecosystems provide sustenance and livelihoods for millions of people around the world. Conserving biodiversity in reef areas is both a social issue and a marine biodiversity priority.
In the face of climate change, Annalisa Bracco, professor in the School of Earth and Atmospheric Sciences at Georgia Institute of Technology, and Lyuba Novi, a postdoctoral researcher, offer a new methodology that could revolutionize how conservationists monitor coral. The researchers applied machine learning tools to study how climate impacts connectivity and biodiversity in the Pacific Ocean’s Coral Triangle — the most diverse and biologically complex marine ecosystem on the planet. Their research, recently published in Nature Communications Biology, overcomes time and resource barriers to contextualize the biodiversity of the Coral Triangle, while offering hope for better monitoring and protection in the future.
“We saw that the biodiversity of the Coral Triangle is incredibly dynamic,” Bracco said. “For a long time, it has been postulated that this is due to sea level change and distribution of land masses, but we are now starting to understand that there is more to the story.”
Connectivity refers to the conditions that allow different ecosystems to exchange genetic material such as eggs, larvae, or the young. Ocean currents spread genetic material and also create the dynamics that allow a body of water — and thus ecosystems — to maintain consistent chemical, biological, and physical properties. If coral larvae are spread to an ecoregion where the conditions are very similar to the original location, the larvae can start a new coral.
Bracco wanted to see how climate, and specifically the El Niño Southern Oscillation (ENSO) in its phases — El Niño, La Niña, and neutral conditions — impacts connectivity in the Coral Triangle. Climate events that move large masses of warm water in the Pacific Ocean bring enormous changes and have been known to exacerbate coral bleaching, in which corals turn white due to environmental stressors and become vulnerable to disease.
“Biologists collect data in situ, which is extremely important,” Bracco said. “But it’s not possible to monitor enormous regions in situ for many years — that would require a constant presence of scuba divers. So, figuring out how different ocean regions and large marine ecosystems are connected over time, especially in terms of foundational species, becomes important.”
Machine Learning for Discovering Connectivity
Years ago, Bracco and collaborators developed a tool, Delta Maps, that uses machine learning to identify “domains,” or regions within any kind of system that share the same dynamic. Bracco initially used it to analyze domains of climate variability in models but also suspected it could be used to study ecoregions in the ocean.
For this study, they used the tool to map out domains of connectivity in the Coral Triangle using 30 years of sea surface temperature data. Sea surface temperatures change in response to ocean currents over scales of weeks and months and across distances of tens of kilometers. These changes are relevant to coral connectivity, so the researchers built their machine learning tool based on this observation, using changes in surface ocean temperature to identify regions connected by currents. They also separated the time periods that they were considering into three categories: El Niño events, La Niña events, and neutral or “normal” times, painting a picture of how connectivity was impacted during major climate events in particular ecoregions.
Novi then applied a ranking system to the different ecoregions they identified. She used rank page centrality, a machine learning tool that was invented to rank webpages on the internet, on top of Delta Maps to identify which coral ecoregions were most strongly connected and able to receive the most coral larvae from other regions. Those regions would be the ones most likely sustain and survive through a bleaching event.
Climate Dynamics and Biodiversity
Bracco and Novi found that climate dynamics have contributed to biodiversity because of the way climate introduces variability to the currents in the equatorial Pacific Ocean. The researchers realized that alternation of El Niño and La Niña events has allowed for enormous genetic exchanges between the Indian and Pacific Oceans and enabled the ecosystems to survive through a variety of different climate situations.
“There is never an identical connection between ecoregions in all ENSO phases,” Bracco said. “In other parts of the world ocean, coral reefs are connected through a fixed, often small, number of ecoregions, and if you eliminate this fixed number of connections by bleaching all connected reefs, you will not be able to rebuild the corals in any of them. But in the Pacific the connections are changing all the time and are so dynamic that soon enough the bleached reef will receive larvae from completely different ecoregions in a different ENSO phase.”
They also concluded that, because of the Coral Triangle’s dynamic climate component, there is more possibility for rebuilding biodiversity there than anywhere else on the planet. And that the evolution of biodiversity in the Coral Triangle is not only linked to landmasses or sea levels but also to the evolution of ENSO through geological times. The researchers found that though ENSO causes coral bleaching, it has helped the Coral Triangle become so rich in biodiversity.
Better Monitoring Opportunities
Because coral reef survival has been designated a priority by the United Nations Sustainable Development Goals, Bracco and Novi’s research is poised to have broad applications. The researchers’ method identified which ecoregions conservationists should try hardest to protect and also the regions that conservationists could expect to have the most luck with protection measures. Their methodology can also help to identify which regions should be monitored more and the ones that could be considered lower priority for now due to the ways they are currently thriving.
“This research opens a lot of possibilities for better monitoring strategies, and especially how to monitor given a limited amount of resources and money,” Bracco said. “As of now, coral monitoring often happens when groups have a limited amount of funding to apply to a very specific localized region. We hope our method can be used to create a better monitoring over larger scales of time and space.”
CITATION: Novi, L., Bracco, A. “Machine learning prediction of connectivity, biodiversity and resilience in the Coral Triangle.” Commun Biol 5, 1359 (2022).
News Contact
Catherine Barzler, Senior Research Writer/Editor
Jan. 03, 2023
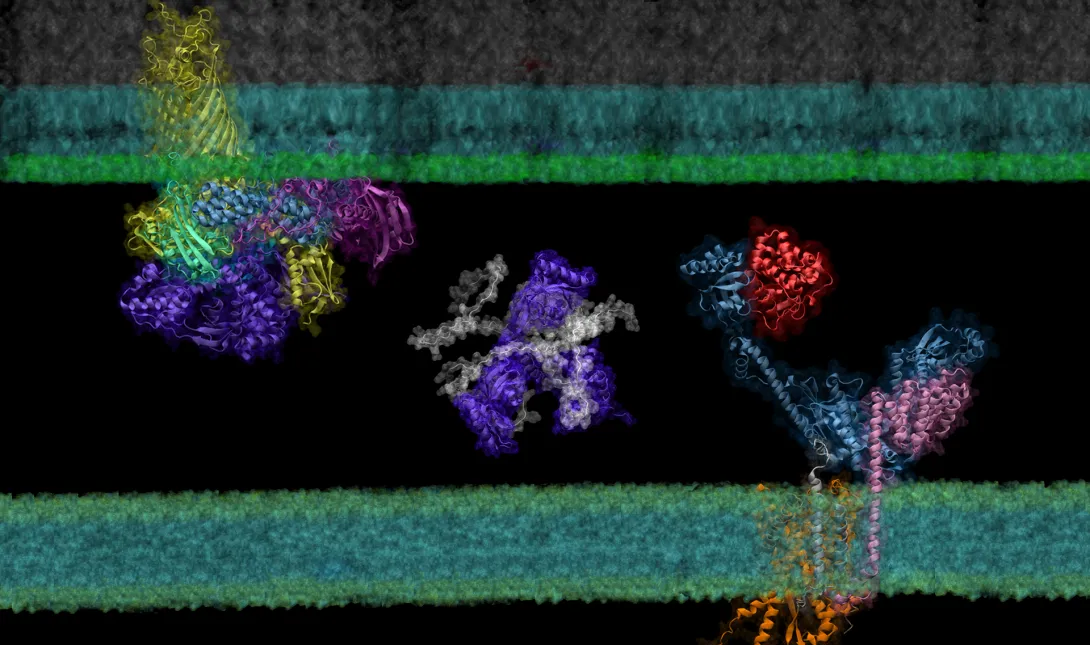
Though it is a cornerstone of virtually every process that occurs in living organisms, the proper folding and transport of biological proteins is a notoriously difficult and time-consuming process to experimentally study.
In a new paper published in eLife, researchers in the School of Biological Sciences and the School of Computer Science have shown that AF2Complex may be able to lend a hand.
Building on the models of DeepMind’s AlphaFold 2, a machine learning tool able to predict the detailed three-dimensional structures of individual proteins, AF2Complex — short for AlphaFold 2 Complex — is a deep learning tool designed to predict the physical interactions of multiple proteins. With these predictions, AF2Complex is able to calculate which proteins are likely to interact with each other to form functional complexes in unprecedented detail.
“We essentially conduct computational experiments that try to figure out the atomic details of supercomplexes (large interacting groups of proteins) important to biological functions,” explained Jeffrey Skolnick, Regents’ Professor and Mary and Maisie Gibson Chair in the School of Biological Sciences, and one of the corresponding authors of the study. With AF2Complex, which was developed last year by the same research team, it’s “like using a computational microscope powered by deep learning and supercomputing.”
In their latest study, the researchers used this ‘computational microscope’ to examine a complicated protein synthesis and transport pathway, hoping to clarify how proteins in the pathway interact to ultimately transport a newly synthesized protein from the interior to the outer membrane of the bacteria — and identify players that experiments might have missed. Insights into this pathway may identify new targets for antibiotic and therapeutic design while providing a foundation for using AF2Complex to computationally expedite this type of biology research as a whole.
Computing complexes
Created by London-based artificial intelligence lab DeepMind, AlphaFold 2 is a deep learning tool able to generate accurate predictions about the three-dimensional structure of single proteins using just their building blocks, amino acids. Taking things a step further, AF2Complex uses these structures to predict the likelihood that proteins are able to interact to form a functional complex, what aspects of each structure are the likely interaction sites, and even what protein complexes are likely to pair up to create even larger functional groups called supercomplexes.
“The successful development of AF2Complex earlier this year makes us believe that this approach has tremendous potential in identifying and characterizing the set of protein-protein interactions important to life,” shared Mu Gao, a senior research scientist at Georgia Tech. “To further convince the broad molecular biology community, we [had to] demonstrate it with a more convincing, high impact application.”
The researchers chose to apply AF2Complex to a pathway in Escherichia coli (E. coli), a model organism in life sciences research commonly used for experimental DNA manipulation and protein production due to its relative simplicity and fast growth.
To demonstrate the tool’s power, the team examined the synthesis and transport of proteins that are essential for exchanging nutrients and responding to environmental stressors: outer membrane proteins, or OMPs for short. These proteins reside on the outermost membrane of gram-negative bacteria, a large family of bacteria characterized by the presence of inner and outer membranes, like E. coli. However, the proteins are created inside the cell and must be transported to their final destinations.
“After more than two decades of experimental studies, researchers have identified some of the protein complexes of key players, but certainly not all of them,” Gao explained. AF2Complex “could enable us to discover some novel and interesting features of the OMP biogenesis pathway that were missed in previous experimental studies.”
New insights
Using the Summit supercomputer at the Oak Ridge National Laboratory, the team, which included computer science undergraduate Davi Nakajima An, put AF2Complex to the test. They compared a few proteins known to be important in the synthesis and transport of OMPs to roughly 1,500 other proteins — all of the known proteins in E. coli’s cell envelope — to see which pairs the tool computed as most likely to interact, and which of those pairs were likely to form supercomplexes.
To determine if AF2Complex’s predictions were correct, the researchers compared the tool’s predictions to known experimental data. “Encouragingly,” said Skolnick, “among the top hits from computational screening, we found previously known interacting partners.” Even within those protein pairs known to interact, AF2Complex was able to highlight structural details of those interactions that explain data from previous experiments, lending additional confidence to the tool’s accuracy.
In addition to known interactions, AF2Complex predicted several unknown pairs. Digging further into these unexpected partners revealed details on what aspects of the pairs might interact to form larger groups of functional proteins, likely active configurations of complexes that have previously eluded experimentalists, and new potential mechanisms for how OMPs are synthesized and transported.
“Since the outer membrane pathway is both vital and unique to gram-negative bacteria, the key proteins involved in this pathway could be novel targets for new antibiotics,” said Skolnick. “As such, our work that provides molecular insights about these new drug targets might be valuable to new therapeutic design.”
Beyond this pathway, the researchers are hopeful that AF2Complex could mean big things for biology research.
“Unlike predicting structures of a single protein sequence, predicting the structural model of a supercomplex can be very complicated, especially when the components or stoichiometry of the complex is unknown,” Gao noted. “In this regard, AF2Complex could be a new computational tool for biologists to conduct trial experiments of different combinations of proteins,” potentially expediting and increasing the efficiency of this type of biology research as a whole.
AF2Complex is an open-source tool available to the public and can be downloaded here.
This work was supported in part by the DOE Office of Science, Office of Biological and Environmental Research (DOE DE-SC0021303) and the Division of General Medical Sciences of the National Institute Health (NIH R35GM118039). DOI: https://doi.org/10.7554
News Contact
Writer: Audra Davidson
Communications Officer
College of Sciences at Georgia Tech
Editor: Jess Hunt-Ralston
Director of Communications
College of Sciences at Georgia Tech
Oct. 25, 2022
During the summer, Duncan Hughes, an Environmental Technology instructor at North Georgia Technical College (NGTC) introduced his students to the web application Virtual Ecological Research Assistant, better known as VERA. It allowed students to construct conceptual models and ecological systems, as well as run interactive model simulations on the brook trout, a species of freshwater fish.
Hughes and his students sought to answer questions about reproduction and food supply, as they worked to add new complexities to the VERA application from different species of trout, circumstances, to changes. According to the Encyclopedia of Life (EOL), an international effort, led by the Smithsonian Institution's National Museum of Natural History, brook trout are found in three types of aquatic environments: rivers, lakes, and marine areas and their living requirements in these environments.
“Originally when we populated the brook trout, we noticed the brown trout shared the same life history and ecological information, but we were able to find enough information from the Encyclopedia of Life to differentiate those species,” said Hughes. “I had my students run through the process of building these components through an instructional-based format by having them manipulate some of the parameters and probabilities.”
VERA was developed by the Design & Intelligence Lab at Georgia Tech in collaboration with EOL. The technology is being used by students as an assisting tool and is publicly accessible. The data being collected from their usage is part of the research conducted at the NSF AI Institute for Adult Learning and Online Education (AI-ALOE).
“Users can jump into our program and conduct ‘what if’ experiments by adjusting simulation parameters. This is our way of providing an accessible and informal learning tool,” said Ashok Goel, director and co-principal Investigator of AI-ALOE and computer science professor at Georgia Tech. “Using VERA as an assessment tool is excellent. These students are using VERA in a way we are not.”
Goel was recently joined by Georgia Tech graduate researcher Andrew Hornback, research scientist Sandeep Kakar, and staff member Daniela Estrada at NGTC to learn more about the work in VERA and challenges Hughes and his students faced while using the application.
“The main struggle is limitation with the EOL and database,” said Hughes. “There are some species that we just can’t find, and sometimes it is glitchy and doesn’t work right away, but it is not insurmountable.”
Another challenge Hughes’ students found was not being able to find what they wanted to complete certain tasks, such as stream and environmental patterns of comparative fish ecosystems.
With that being known, AI-ALOE is working to address these issues and more to build and cater to specific student and teacher needs. At this time, the Design & Intelligence Laboratory is in the process of expanding VERA in the capability of its on-demand agent-based simulation generator, which would enable users to divide components into separate habitats.
“It was very interesting to see the results because antidotally through much research we were able to set up all these relationships and let them run the model, and the results were exactly what we would have hypothesized what they would be given those perimeters,” said Hughes.
The technical college has plans to introduce VERA to another classroom this semester held by Natural Resource Management instructor, Kevin Peyton.
About VERA
Interested in trying out VERA? Create an account at https://vera.cc.gatech.edu/. You can also find VERA’s user guide as well as a step-by-step tutorial at http://epi.vera.cc.gatech.edu/docs/exercise.
About AI-ALOE
The NSF AI Institute for Adult Learning and Online Education (AI-ALOE) is developing an AI-based transformative model for online adult learning through research and data collection.
About NGTC
North Georgia Technical College is a residential, public, multi-campus institution of higher education serving the workforce development needs of Northeast Georgia and part of the Technical College System of Georgia.
News Contact
Breon Martin
AI Communications Officer
breon.martin@gatech.edu
Pagination
- Previous page
- 10 Page 10